Introduction:
Parhelia Biosciences is excited to announce the development of an automated Gram staining protocol for microbial samples using the Omni-Stainer™ S12 Automation Bundle. We decided to investigate the applications of this technology in microbiology and biomanufacturing after it was originally designed for automated precision staining in mammalian tissues.
One of the key advantages of Parhelia Omni-Stainer™ is its ease of use and affordability in comparison to other autostainers on the market. Another significant benefit is flexibility, which allowed our R&D team to develop and verify the Gram staining protocol in a matter of days.
Most importantly, when compared to manual slide staining, the Omni-Stainer™ provides increased reproducibility, precision, safety, and reagent savings, once common staining protocols for microbial samples are developed and tested. In fact, Omni-Stainer™ capillary gap staining uses 50-75% less reagents than open-air staining methods and procedures improved staining quality and uniformity.
Parhelia Biosciences is excited to offer this innovative technology to researchers and looks forward to seeing the exciting results it will enable.
This white paper describes the development and testing of one very common staining protocol, the Gram Staining Protocol.
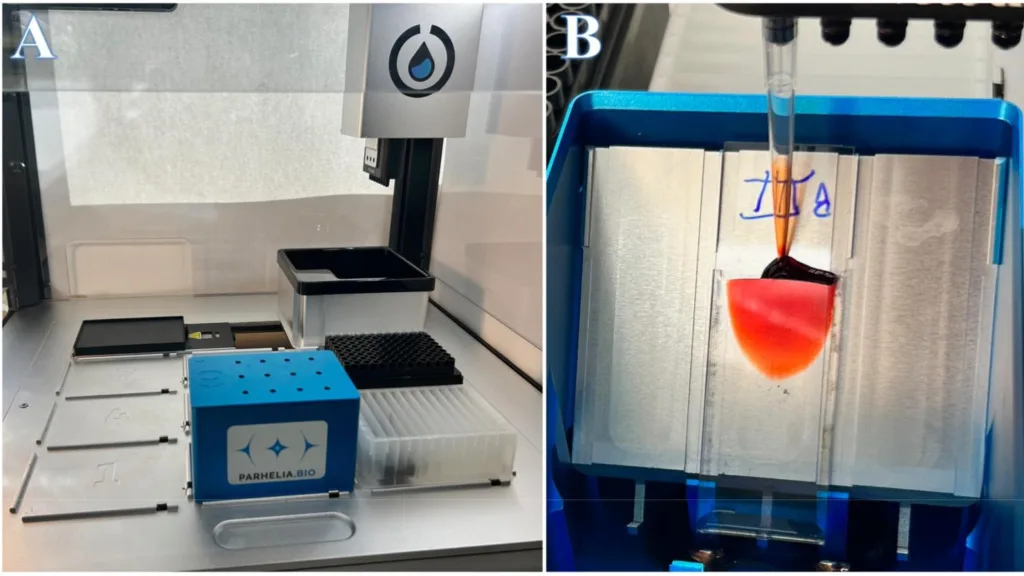
Omni-Stainer S12 Automation Bundle shown with lid on, in the Opentrons OT-2 robotic liquid handler.
Figure 1B, Closeup of Capillary Laminar Flow.
Counterstaining step from a successful Gram staining protocol run, showing the addition of 100 μl Safranin Red solution.

Animation from Gram staining protocol run, showing the addition of 100 μl Crystal Violet solution.
Background:
Gram staining, one of the most widely used staining methods in microbiology, distinguishes different types of bacteria – making it an excellent proof of concept for microbial applications in automated staining with the Omni-Stainer™. Traditional manual reagent application and washing steps are error-prone and time-consuming. The Omni-Stainer S12 automates the process by using capillary action and reagent displacement to produce uniform and reproducible results with less wash water and fewer reagents.
Methods – Experimental Design:
We prepared replicate microscopy slide smears from pure cultures (e.g., E. coli DH5-α) and environmental samples (e.g., dental plaque, soil) by heat fixation. Then, we Gram-stained slides by hand using a common method outlined by Bio-Rad Laboratories, in parallel, replicate smears were Gram-stained with the Omni-Stainer™ S12 system on the Opentrons OT-2 robotic liquid handler (Figure 2). Finally, we then mounted coverslips on the slides and imaged them with an optical light microscope (Figure 4).
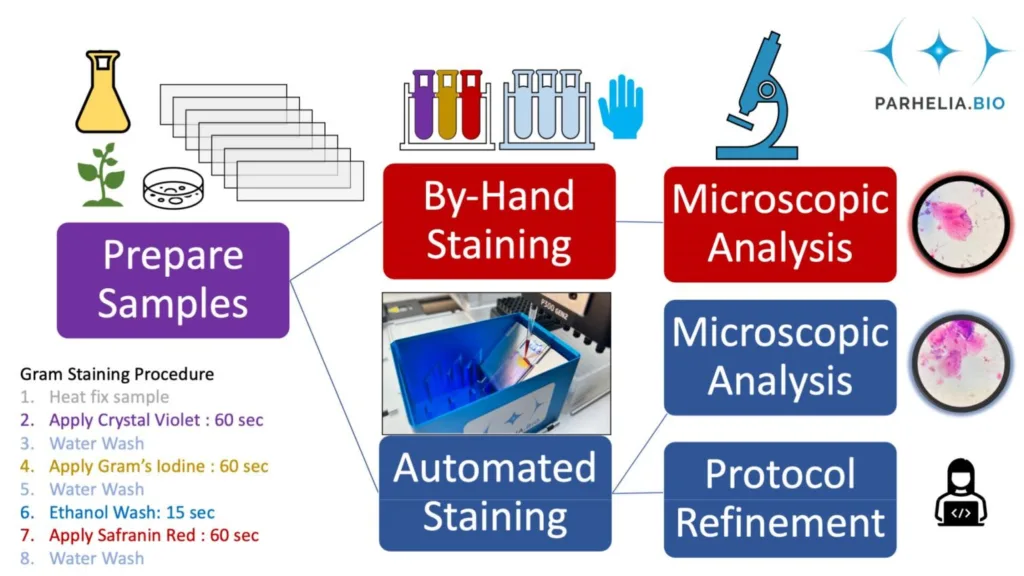
In the Omni-Stainer™ system, reagents flow over the cells, which are heat fixed to the microscope slides. Therefore we set out to verify that the microbial cells don’t wash off the slide, since tissues and microbial samples are different – and to check this for a diverse set of microbial samples.
Methods – Description of the autostaining technology:
The Parhelia Omni-Stainer™ S12 system is compatible with essentially any robotic liquid handler that utilizes pipette tips or can be used manually with a micropipette. This new automated gram staining protocol was created for the Opentrons OT-2 robot, which is significantly less expensive than other similar robots on the market. The Omni-Stainer S12 slide staining module has a standard SBS footprint (the footprint of a 96-well plate) and can stain 12 slides at a time, making it more versatile and compact than stand-alone automated cell staining technologies.
Traditional open-air gram staining involves flooding the slide with a specific stain (e.g., crystal violet), washing it off in water, and then applying the next stain. The length of time each step of the protocol takes is critical, and a difference of 5 seconds for a step in the protocol will change the results. It can be difficult for us humans to keep precise track of these steps while attempting to perform other tasks or to keep track of these steps for a large number of samples at once.
The Omni-StainerTM S12 Automation Bundle, on the other hand, operates through capillary action and the displacement of the staining or wash reagent by application of another reagent. We optimized the Omni-StainerTM S12 system to keep staining reagents on the slide until displaced, even after extremely long incubations. With automation, users can click a few buttons and walk away confident that incubation times are being precisely followed. Displacement via laminar flow ensures samples are stained evenly.
The capillary laminar flow is clearly demonstrated below (Figure 3) by the red solution flowing into and displacing the water.
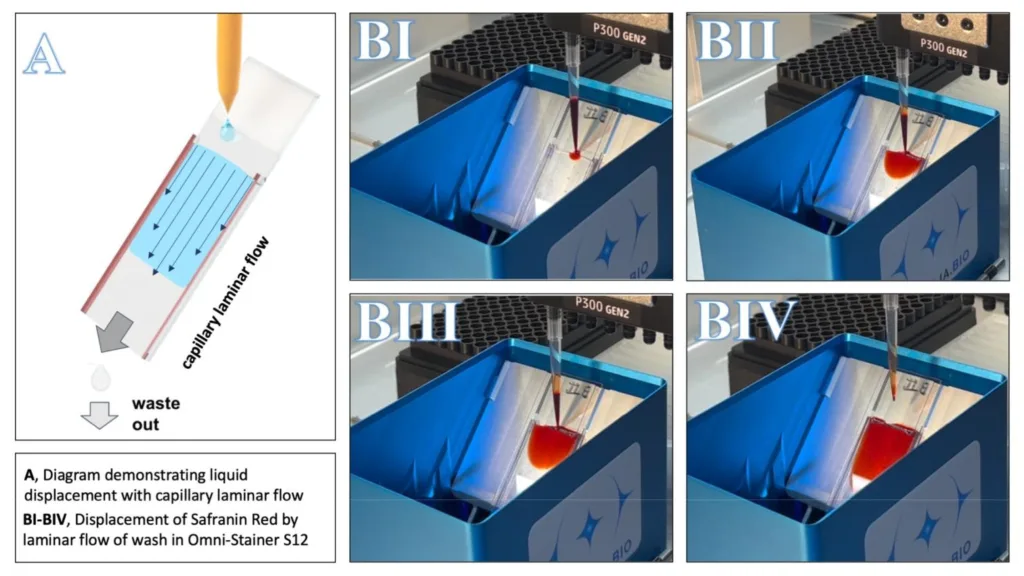
Figure 3A shows the principles utilized by the Omni-Stainer. The velocity profile needed for laminar flow is shown with thin vector arrows. Figure 3B(I-IV) shows a time course (approx. 2 seconds) of the addition of 100 μl Safranin red solution, the counterstaining step from a successful Gram staining protocol run.
Results:
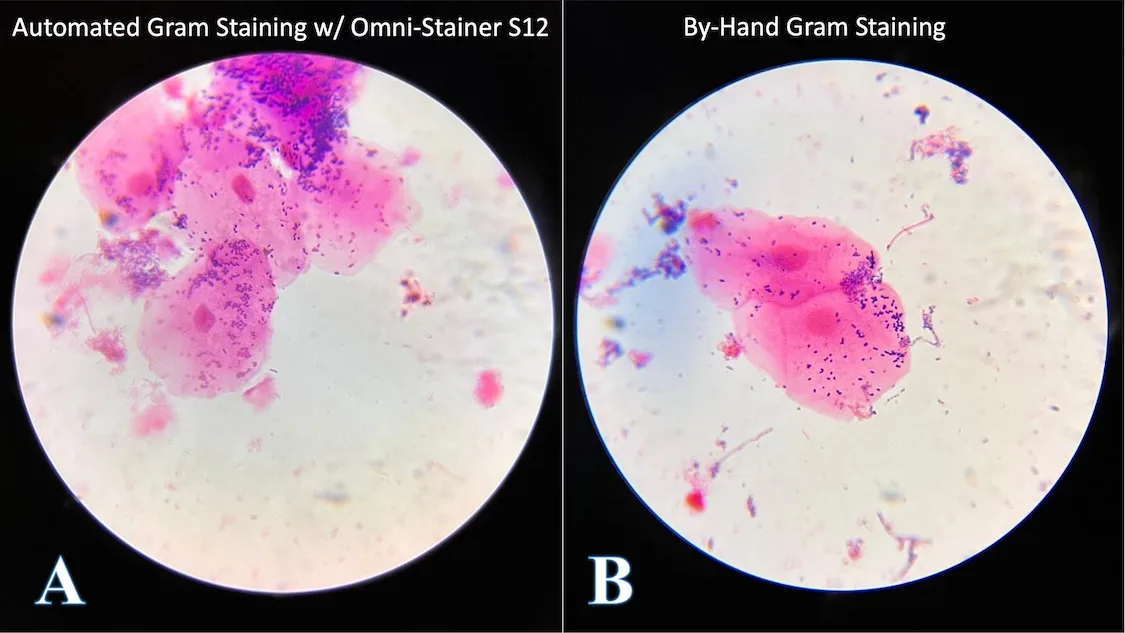
We collected human oral microbiome samples and stained them using our new Automated Gram Staining Protocol on the Omni-Stainer™ S12 system, run by an Opentrons OT-2 robotic liquid handler (Figure 4A). The results were compared to manual stains performed by an expert microbiologist (Figure 4B). The images were captured using an brightfield microscope using an 100x oil immersion objective.
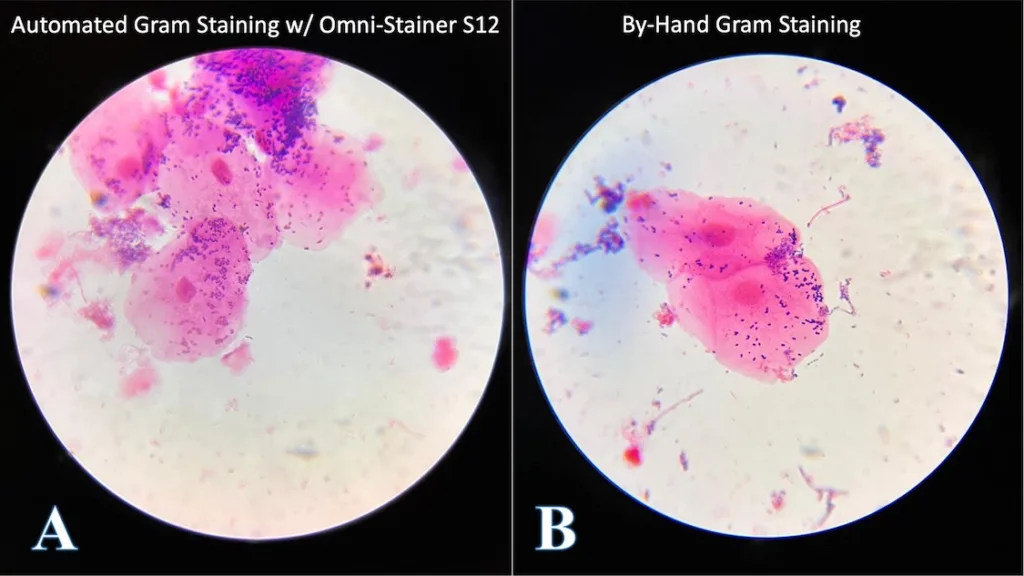
The large pink eukaryotic cells are mouth epithelial cells, and the small purple (purple = Gram-positive) paired circles are most likely a type of Streptococcus, a common facultative anaerobe in the human microbiome. Despite that only one sample type is shown, we tested our protocol on variety of microbial samples, including pure cultures of bacteria and yeast – as well as soil extractions – and the results were consistent.
The stained replicate smears had no qualitative differences in microscopic appearance.
Automated Gram Staining Applications:
Our automated Gram staining technology has numerous potential applications in a variety of industries and sectors, including microbiology research, biomanufacturing, and biotechnology. Here are a few examples:
- In microbiology, the technology could be used to accelerate and improve the consistency of bacterial and microbial sample analysis, enabling researchers to process more samples in less time and with fewer errors.
- In biomanufacturing, the technology could be used to streamline and improve the efficiency of quality control processes, helping to ensure that products are free of contaminants and meet regulatory standards.
- In biotechnology research, the technology could be used to help identify and characterize various microorganisms – such as antibiotic susceptibility testing to discover new antibiotics.
Challenges and Limitations:
While there are many potential benefits to utilizing the automated Gram staining we developed – including its affordability in comparison to other automated cell staining technologies – we recognize that there may be some challenges and limitations.
The upfront cost of purchasing the necessary equipment, which may be prohibitive for some laboratories or research groups is one potential challenge. However, in medium- to high-throughput applications, the use of substantially smaller volumes of reagents and reduced waste production can quickly offset this initial investment.
Another potential challenge is that full automation in some labs may necessitate specialized training, which could be a barrier to adoption for some users. We have step-by-step instructions with pictures and videos. Our support team, Opentrons support, and the community of active Omni-Stainer users are happy to answer specific questions.
Finally, the technology may not be suitable for all types of microbial samples, and additional user acceptance testing research may be required to optimize the protocol for various types of samples. Since capillary action requires a narrow space, a defined volume exists between the microscope slide and the S12 Cover Tile, meaning any sample that is thicker than 50 μm would not be stained efficiently. This was observed in one sample that was deliberately made tall during our testing. This constraint may limit the use of capillary gap staining when studying thick biofilms or microbial interaction with thick surfaces. However, these situations are rare, and the vast majority of bacterial and fungal microscope samples are unaffected.
Future Research and Development:
We see several areas of future research and development related to the automated Gram staining technology, as well as numerous microbiology applications for automated cell staining beyond Gram staining, including:
- Ongoing optimization of the protocol for different types of microbial samples, to improve the staining process’s effectiveness and accuracy.
- Integration of the Omni-Stainer technology with other automated laboratory equipment and systems, such as robotic microscopes or image recognition and data analysis software.
- We believe that the Gram Staining Protocol, either alone or in conjunction with other technologies, has great potential for identifying and tracking changes in bacterial populations over time and monitoring the effectiveness of antimicrobial treatments.
Beyond Gram Staining, we see many potential applications for automated cell staining technology in microbial assays. Such as rRNA-FISH (Fluorescence In Situ Hybridization), DNA-FISH, and immunofluorescence for microbes. Researchers can use these tools to visualize phylogenetic and transcriptomic data spatially. This resolution empowers researchers in many ways, including monitoring and engineering strains for changes that affect critical outcomes such as:
- Antibiotic susceptibility
- Contagiousness
- Toxicity and mutagenicity
- Ecological Fitness
- Trait Development
- Protein expression
- Plasmid copy number production
- Fermentation efficiency
We have previously published complex staining protocols with specific heating and cooling requirements in mammalian tissue samples using Parhelia Bio’s Omni-Stainer™ S12/C12 system in conjunction with the Thermal Sheath and a temperature module.
We have already developed and successfully deployed FISH (Fluorescence In Situ Hybridization), RNA in situ hybridization, and immunofluorescence assays:
- Co-detection by indexing (CODEX)
- Immunohistochemistry (IHC)
- Opal Multiplex IHC Assay
- Hematoxylin & Eosin + Formalin Fixation and Paraffin Embedding (H&E + FFPE) Antigen retrieval.
In addition to onboarding new assays, we want to put relevant autostaining protocols developed for tissue samples to the test on various types of microbial samples, and see if there is anything else we should tweak before recommending these autostaining protocols for microbial applications.
Conclusion:
This Gram Staining Protocol is ready to be used for microbial samples.
Soon, the Opentrons protocol should be available on the Parhelia StainWorksTM library. Additionally, while not explicitly tested for Gram staining, hand-pipetting with the Omni-Stainer will work well based on prior data and experience. However, as with any new laboratory method, additional user acceptance testing is recommended.
The automation and elegance of Parhelia Biosciences’ microbial cell staining technology is an exciting advance in the field of microbiology and biomanufacturing. When compared to traditional open-air staining, our technology provides greater consistency, minimizes waste, and reduces mess by automating the staining process. Plus, our Omni-Stainer system is 20x cheaper than the market-leading autostaining solution – with bonus reagent savings.
In our experiments, the traditional method of Gram staining used around 500 ml of water, while the automated protocol used only 0.5 ml of water per sample.
A roughly, 1000x reduction in wash water that is contaminated with staining reagents!
While the reagent cost for Gram staining (<$1/ml) is negligible when compared to other staining reagents such as IHC (>$300/ml) the reduced waste volumes may result in substantially lower disposal and environmental costs.
Our technology has potential applications in a variety of industries and sectors, including microbiology research, biomanufacturing, and biotechnology. However, we recognize that some challenges and limitations must be addressed in order to fully realize the potential of the technology, including the upfront cost of purchasing the necessary equipment, the need for specialized training, and the potential for limited compatibility with certain types of samples.
We see numerous future R&D opportunities in automated Gram staining and other microbial applications. Including sample optimization and integration of our technology with other automated laboratory equipment and systems. We believe that automated Gram staining technology has the potential to significantly improve the efficiency and accuracy of research processes in a variety of fields, while also providing significant cost savings when compared to other automated cell staining technologies.
While Gram Staining is still widely used in many areas, the more significant advantage of our technology is realized when reagents are scarce and the staining protocols are highly multiplexed and/or complicated.
The demonstrable success of using Parhelia Bio’s technologies for traditional microbial research workflows, such as Gram staining, supports the case for many future microbial applications for Parhelia Bio at the forefront of science and biotechnology!